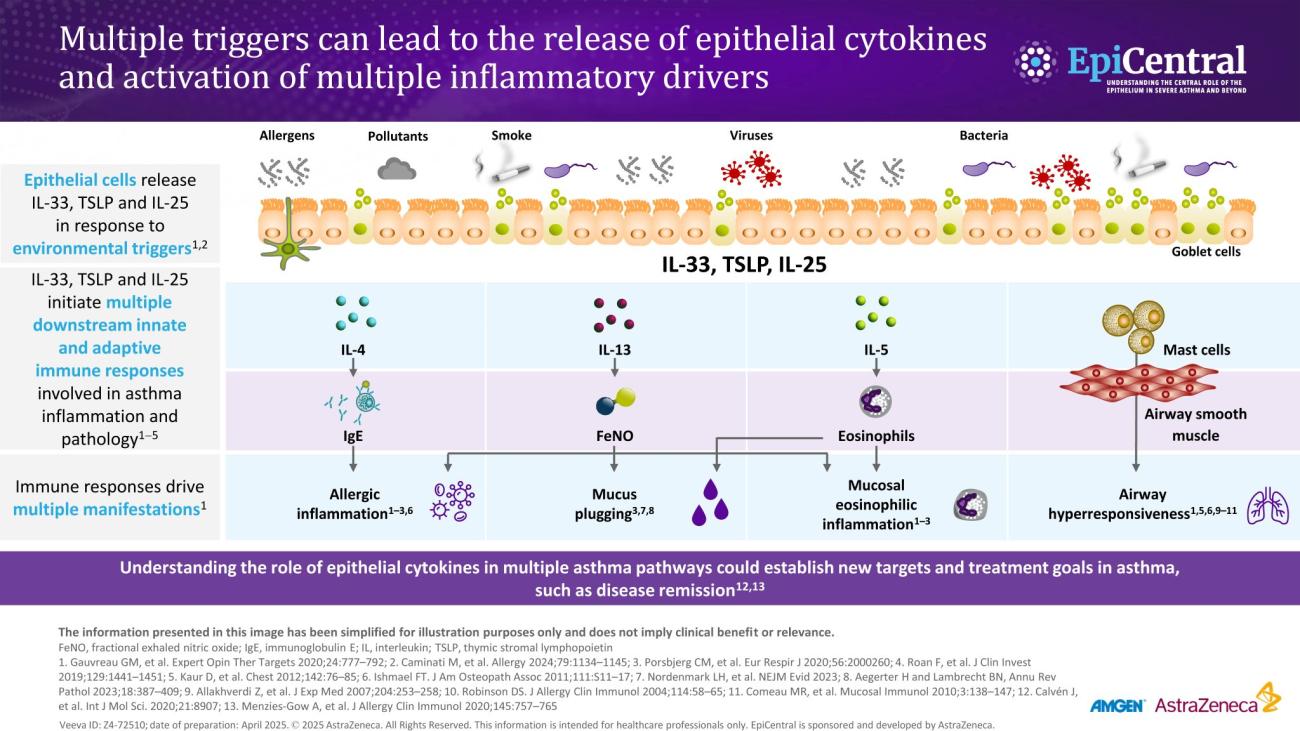
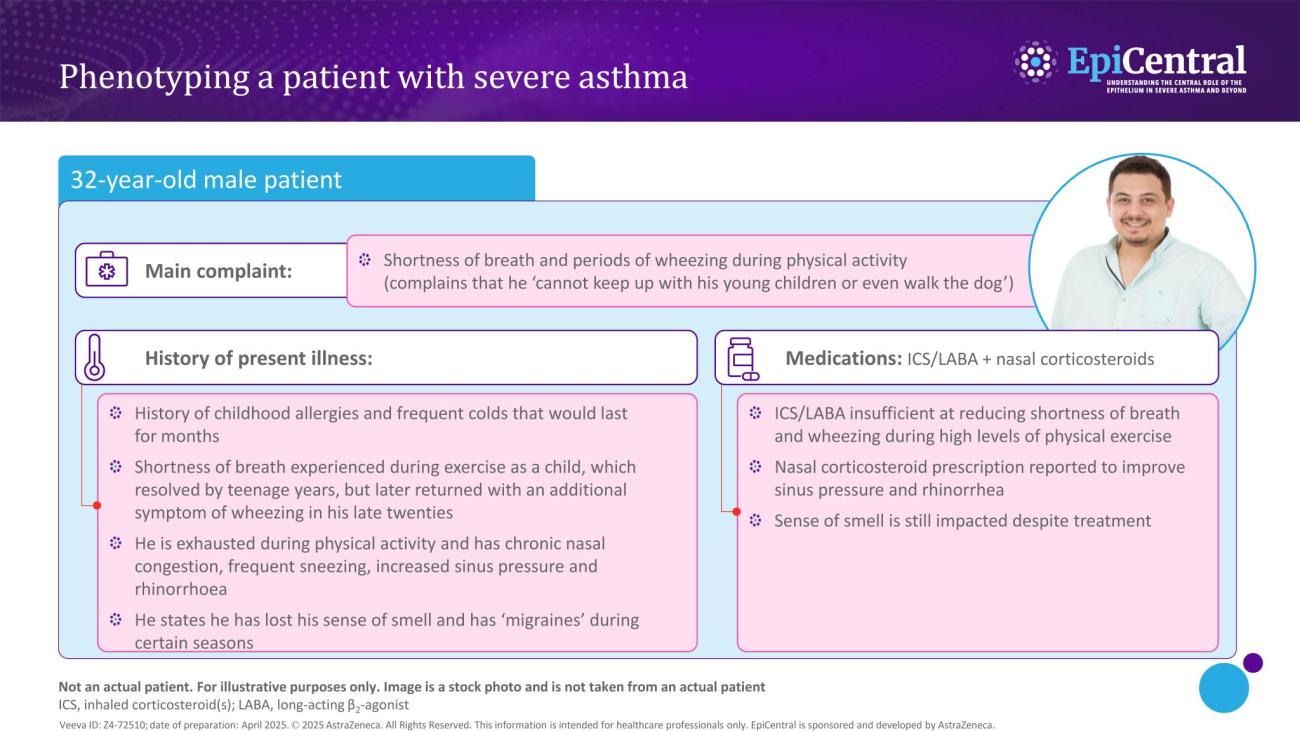
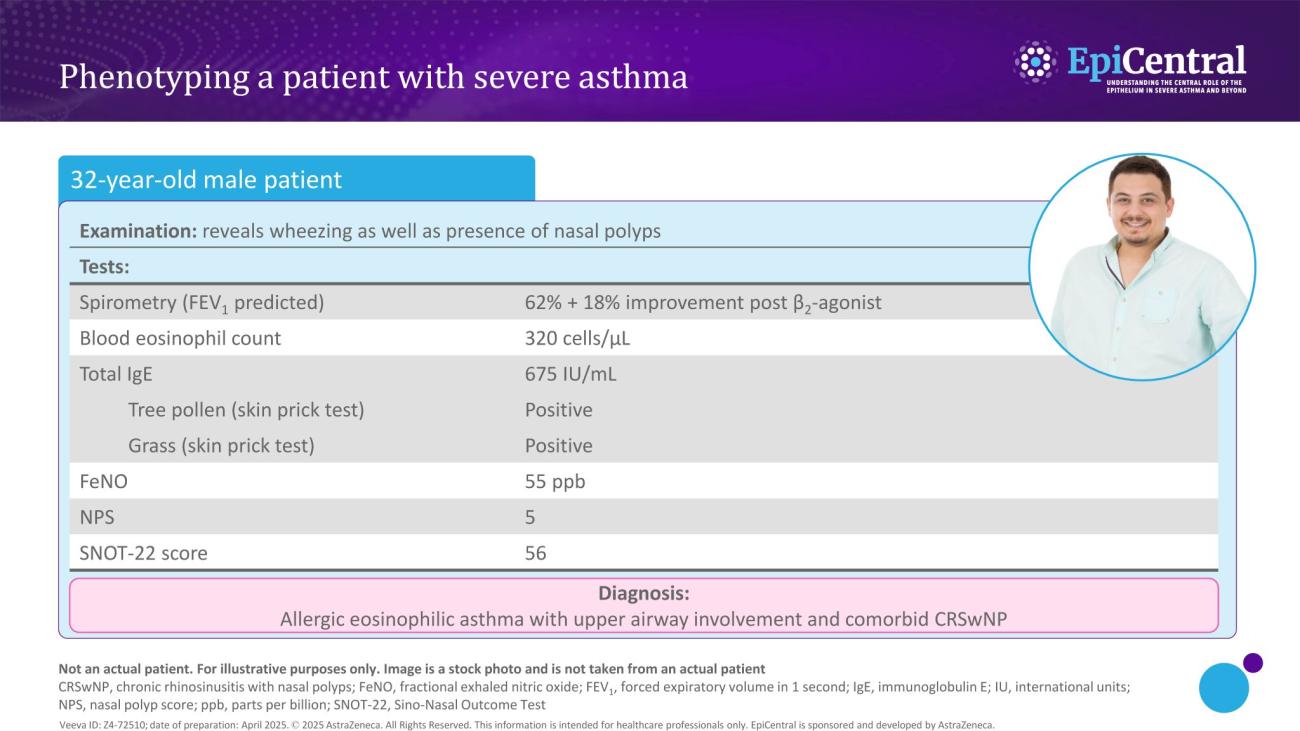
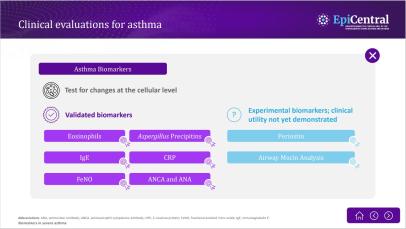
Inflammation in severe asthma
Asthma-associated inflammation is complex and heterogeneous,1–4 and numerous cell types, mediators and downstream immune pathways are involved.1,3–6 Multiple inflammatory endotypes have been characterised – allergic and eosinophilic inflammation, to name a few.5